Visualize pcap file data
One nice day I was given orders to produce network usage statistics to find eventual burst in the network stream. I faced two problems with this task. The network monitoring software was graphing the network flow only every minute which was too coarse and the interface was connected to a switch I had no control over at all, A mirror port had to be ruled out. The assignment was to collect data for a week and then look at the numbers.
I was unsure how to go about it in the first place. To not fall behind I ran tcpdump on the hosts in question until the week was over. At that point in time I had no idea how to process the pcap dump data and I had not the faintest clue how to present it at the end of the week. After a lot of searching I finally came across a nifty feature in tshark allowing me to aggregate bandwidth on a per second basis. Below is a short recipe how to create graphs from captured network traffic.
Prerequisites
- A capture file the wireshark suite understands. E.g. pcap or Solaris snoop among others.
- tshark
- R
- ImageMagick's montage [optional]
Howto
Aggregate traffic with tshark
To properly graph the data tshark needs to generate statistic on a per second basis. The below command will achive this.
tshark -q -z 'io,stat,1' -r <PcapFile> > <StatisticsFile>
The output is looking something like the excerpt below.
<<<<<<< <StatisticsFile> ======= Time |frames| bytes 000.000-001.000 62 5578 001.000-002.000 62 5386 002.000-003.000 62 5692 003.000-004.000 62 5968 004.000-005.000 62 5428 005.000-006.000 62 5838 006.000-007.000 62 5912
The only problem with the output above is that the time is relative to the start of the pcap file. Before passing the data to R it has to be properly massaged.
Produce a graph with R
Finally to produce the graph in PDF or PNG format the below R script is being used.
#!/usr/bin/Rscript
## ----------------------------------------------------------------------------
## Globals for reading the data file
## ----------------------------------------------------------------------------
skip.header <- 1 # how many lines to skip including the header row
comment.char <- "=" # skip lines with <char> in it
## ----------------------------------------------------------------------------
## Don't touch below unless you know what you are doing
## ----------------------------------------------------------------------------
col.names <- c( "time", "frames", "bytes" )
args <- commandArgs( trailingOnly = TRUE )
number.graphs <- length( args )
for ( d in 1:length( args ) ) {
file <- args[[d]]
# get date and time from file name and convert to a time object
date.time <- as.POSIXlt(
gsub(
".*([0-9]{4}-[0-9]{2}-[0-9]{2})_([0-9]{2})-([0-9]{2}).*",
"\\1 \\2:\\3:00",
file,
perl=T
)
)
# read the data
traffic <- read.table( file=file,
header=F,
col.names=col.names,
skip=skip.header,
comment.char="="
)
# massage the data a bit
traffic$kbits <- ( traffic$bytes * 8 ) / 1024
traffic$frames <- NULL
traffic$bytes <- NULL
traffic$time <- as.numeric(
gsub("-.*", "", traffic$time, perl = T )
) + date.time
# calculate max an avg
traffic.max <- round( max( traffic$kbits ), digits = 2 )
traffic.avg <- round( mean( traffic$kbits ), digits = 2 )
# prepare the graph
sub.title <- paste( "Max:", traffic.max, "Kbit/s; Avg:", traffic.avg, "Kbit/s" )
names( traffic )
# output as pdf and png
pdf( paste( file, ".pdf", sep = "" ) )
plot( traffic, type="h", main=file, sub=sub.title, xlab="Time", ylab="Kbit/s" )
png( paste( file, ".png", sep = "" ) )
plot( traffic, type="h", main=file, sub=sub.title, xlab="Time", ylab="Kbit/s" )
}
To run the script issue the following command assuming the above script is called tshark-graph.R.
Rscript tshark-graph.R *ts
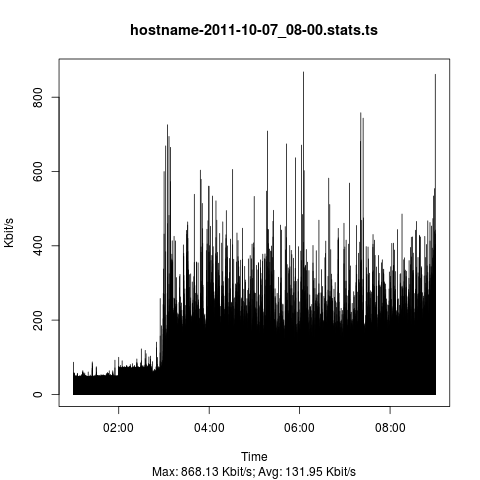
Resulting graph
There are sexier graphs out there but from a functional standpoint it does the job.
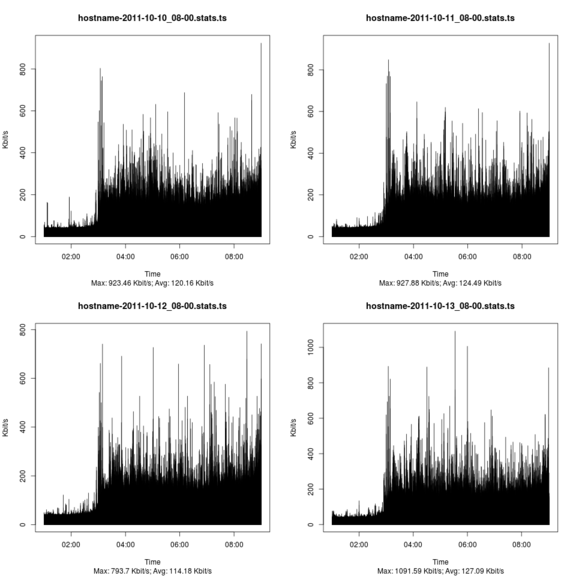
Combining graphs
R is fully capabale of creating a collection of graphs from a bunch of files but personally I think it's a lot more involved than simply using ImageMagick's montage command.
montage -geometry <Width>x<Height> <GraphFiles> <OutputGraph>
Yields a similar graph like the one below (intentionally downsampled to fit page)